本周课程要点: - 含有单独隐藏层的神经网络 - 参数初始化 - 使用前向传播进行预测 - 以及在梯度下降时使用反向传播中 - 涉及的导数计算
课程笔记
Neural Network Overview
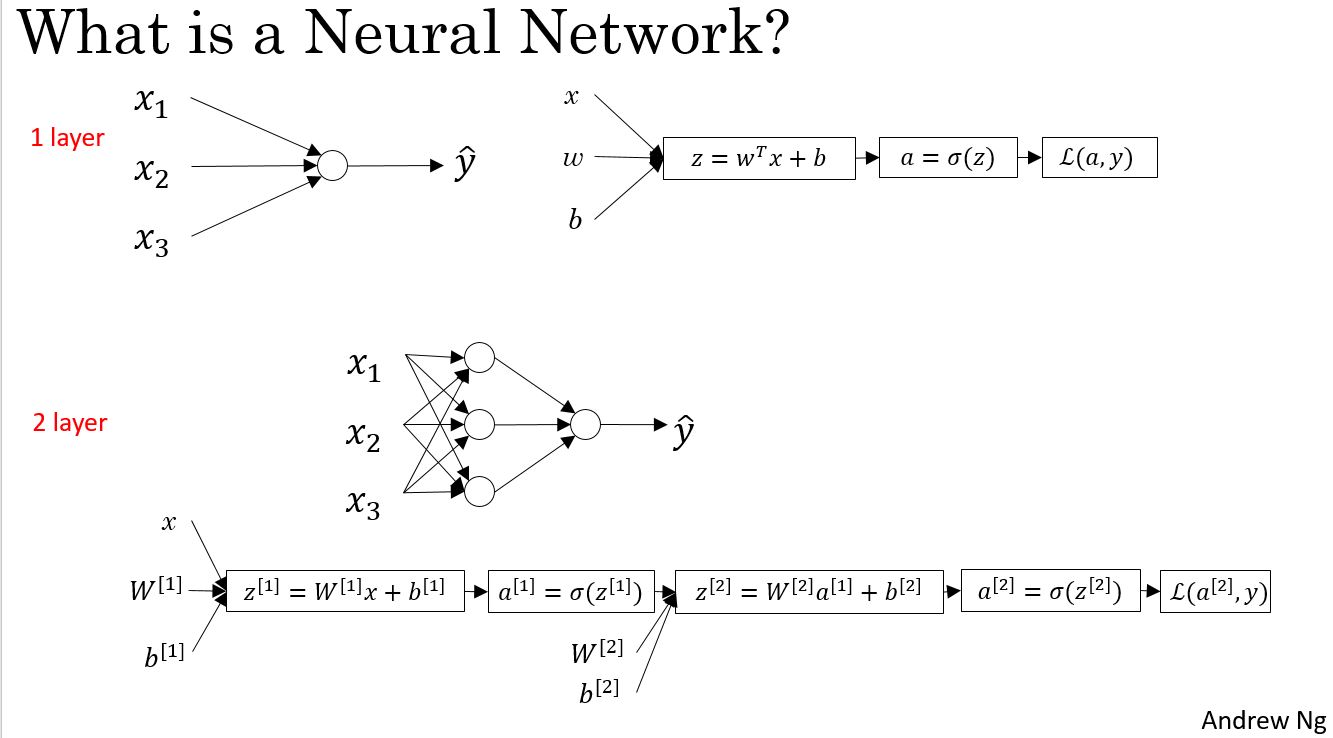
Neural Network Represention
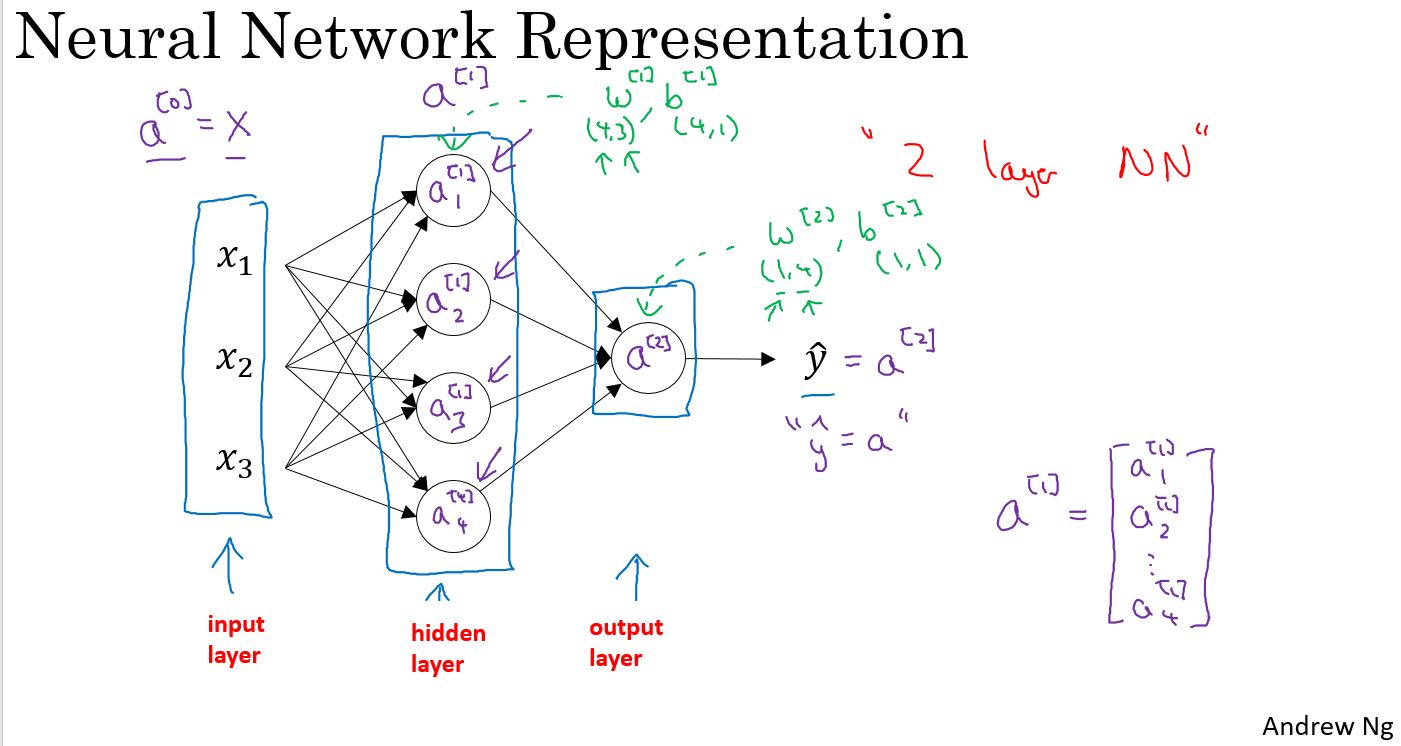
Neural Network Computing and Vectorizing
对于一层的神经网络,每个网络节点都会进行如下两个运算:
$ z = w^T x + b $
$ = a = (z) $
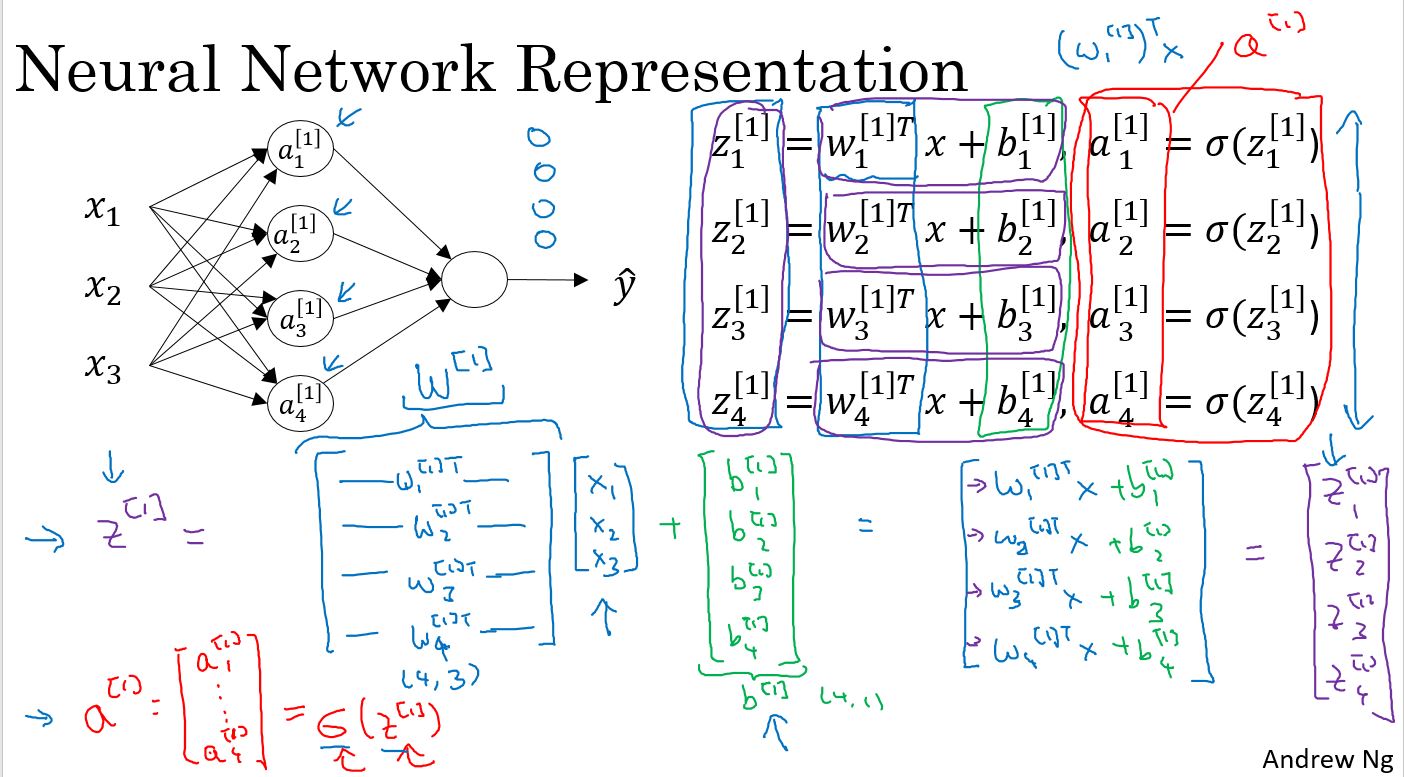
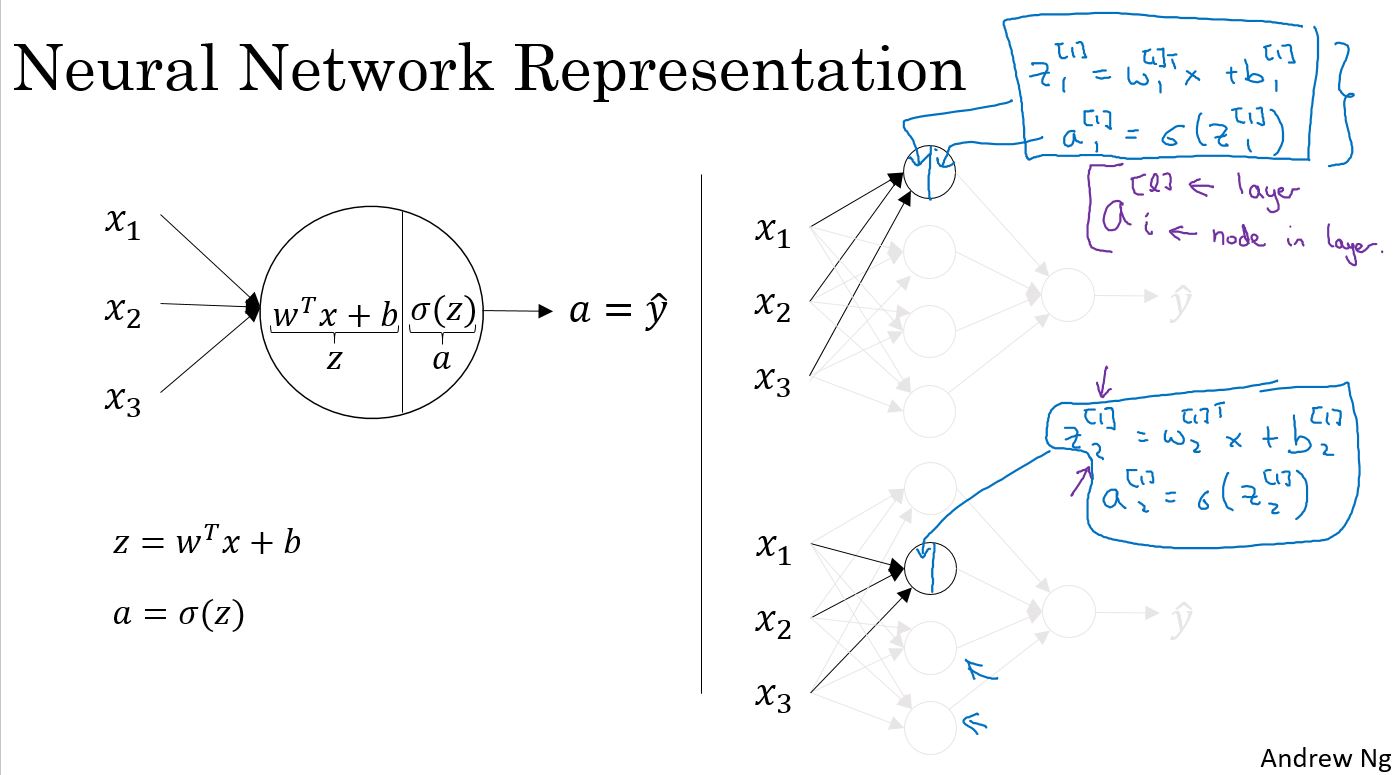
上图中的手写部分,是把输入和参数矢量化。
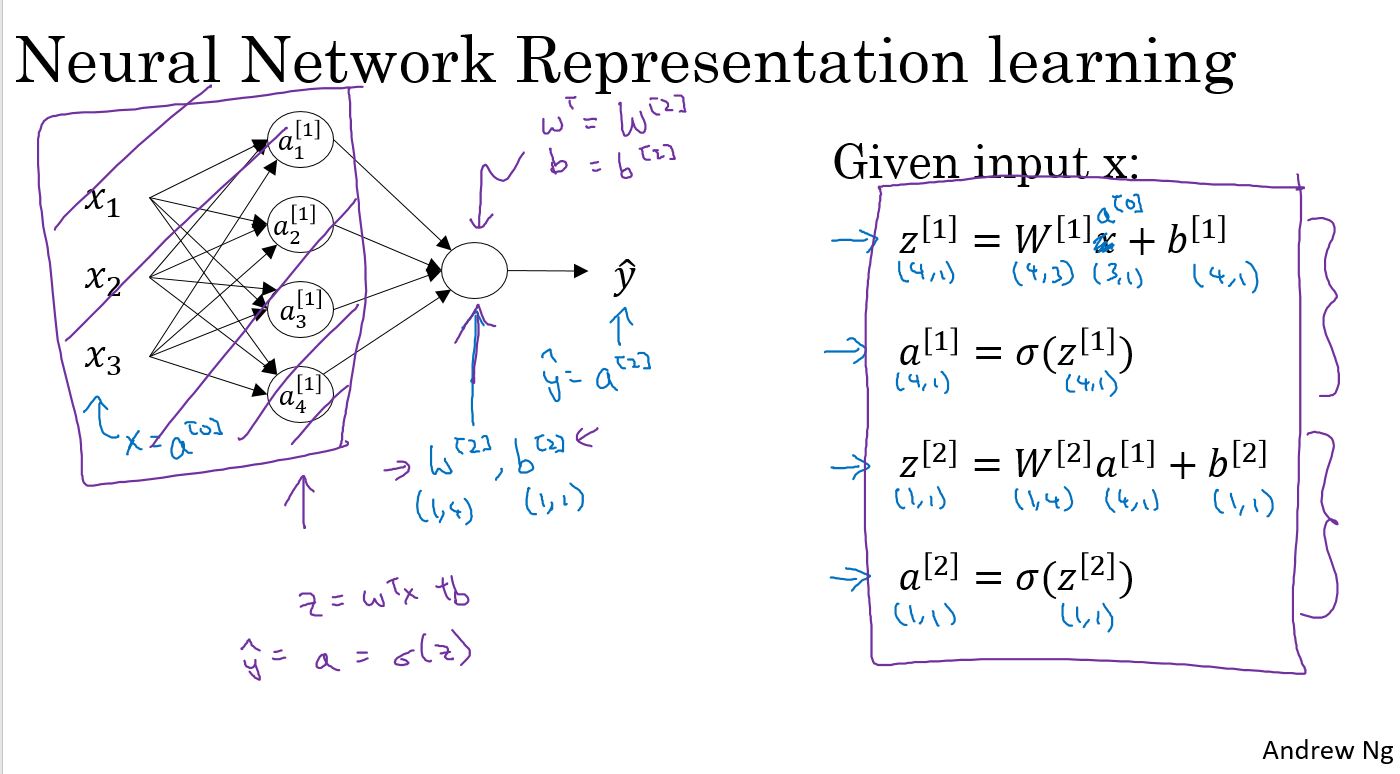
上图中是一个两层网络,第一层是4个节点,第二层1个节点,通过矢量化,并且把输入作为\(a^{[0]}\),可以归结为右侧的4个公式。
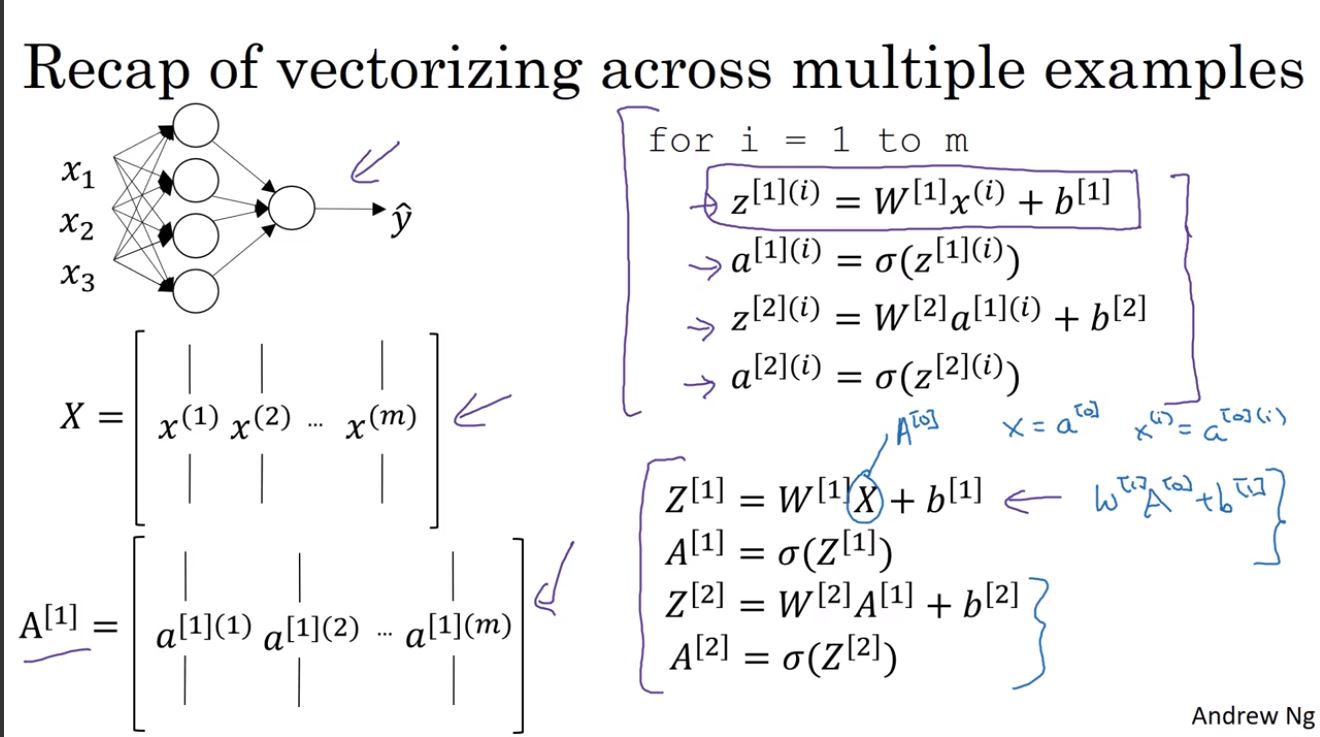
激活函数
week2中用到的只有一个激活函数:sigmoid函数,除此之外还有其他激活函数。
- sigmoid函数 \[ sigmoid(z) =\frac{1}{1+e^{-z}} \]
- tanh函数 \[ tanh(z) =\frac{e^z-e^{-z}}{e^z+e^{-z}} \]
- ReLU \[ f(x) = \begin{cases} x, & \text{if $x$ >= 0} \\ 0, & \text{if $x$ < 0} \end{cases} \]
- LReLU \[ f(x) = \begin{cases} x, & \text{if $x$ >= 0} \\ \alpha x, & \text{if $x$ < 0} \end{cases} \]
曲线分别为:
Why do you need non-linear activation functions?
为什么要使用激活函数?
如果你使用“线性激活函数”或者叫“恒等激活函数”,那么神经网络的输出仅仅是输入函数的线性变化。深度网络会有很多层,很多隐藏层的神经网络。如果使用线性激活函数 或者说 没有使用激活函数,那么无论神经网络有多少层,它所做的仅仅是计算线性激活函数,这还不如去除所有隐藏层。请记得:线性的隐藏层没有任何用处,因为两个线性函数的组合,仍然是线性函数,除非在这里引入非线性函数,否则无论神经网络模型包含多少隐藏层,都无法实现更有趣的功能。
神经网络的梯度下降
对于一个2层的神经网络,使用梯度下降法计算的方式为:
- Paramters : \[ W^{[1]},\,\,\,b^{[1]},\,\,\,W^{[2]},\,\,\,b^{[2]} \]
其维度分别为:
\[ dims:(n^{[1]},\,\,\,n^{[0]}),\,\,\,(n^{[1]},1),\,\,\,(n^{[2]},n^{[1]}),\,\,\,(n^{[2]},1) \]
- Cost Function : \[ J(W^{[1]},b^{[1]},W^{[2]},b^{[2]}) = \frac{1}{m} \sum_{i=1}^m \mathcal{L}(a^{(i)}, y^{(i)}) \]
- Gradient descent : \[ \begin{array}{l} \text{Repeat \{} \\ \,\,\,\,\,\,\,\,Compute\,\, predicts(\hat{y}_{(i)},i=1,2,\cdots,m) \\ \,\,\,\,\,\,\,\, dW^{[1]} = \frac{d\mathcal{L}}{dW^{[1]}},db^{[1]} = \frac{d\mathcal{L}}{db^{[1]}} \\ \,\,\,\,\,\,\,\, dW^{[2]} = \frac{d\mathcal{L}}{dW^{[2]}},db^{[2]} = \frac{d\mathcal{L}}{db^{[2]}} \\ \,\,\,\,\,\,\,\,W^{[1]} = W^{[1]} - \alpha * dW^{[1]} \\ \,\,\,\,\,\,\,\,b^{[1]} = b^{[1]} - \alpha * db^{[1]} \\ \,\,\,\,\,\,\,\,W^{[2]} = W^{[2]} - \alpha * dW^{[2]} \\ \,\,\,\,\,\,\,\,b^{[2]} = b^{[2]} - \alpha * db^{[2]} \\ \text{\}} \end{array} \]
进行梯度计算的公式
前向计算
$ Z^{[1]} = {W^{[1]}} X + b^{[1]} $
$ A^{[1]} = g{[1]}(Z{[1]}) $
$ Z^{[2]} = {W^{[2]}} X + b^{[2]} $
$ A^{[2]} = g{[2]}(Z{[2]}) $
对二分类问题,\(g^{[2]}\)依然使用\(sigmoid\)函数。
反向传播
$ dZ^{[2]} = A^{[2]} - Y $
$ dW^{[2]} = dZ^{[2]} {A{[1]}}{T} $
$ db^{[2]} = np.sum(dZ^{[2]},axis=1) $
$ dZ^{[1]} = (W{[2]})T dZ^{[2]} (g{[1]})'(Z{[1]}) $
$ dW^{[1]} = dZ^{[1]} X^{T} $
$ db^{[1]} = np.sum(dZ^{[1]},axis=1) $
反向传播的公式推导
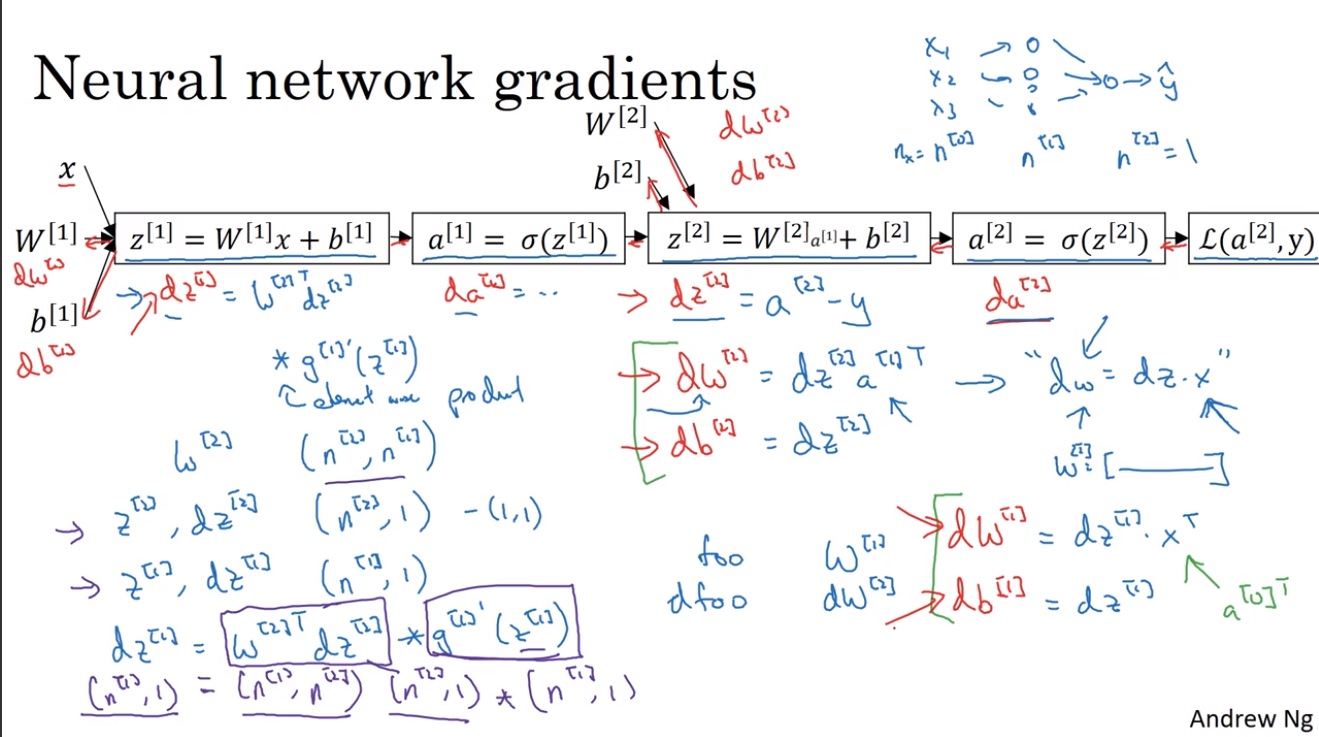
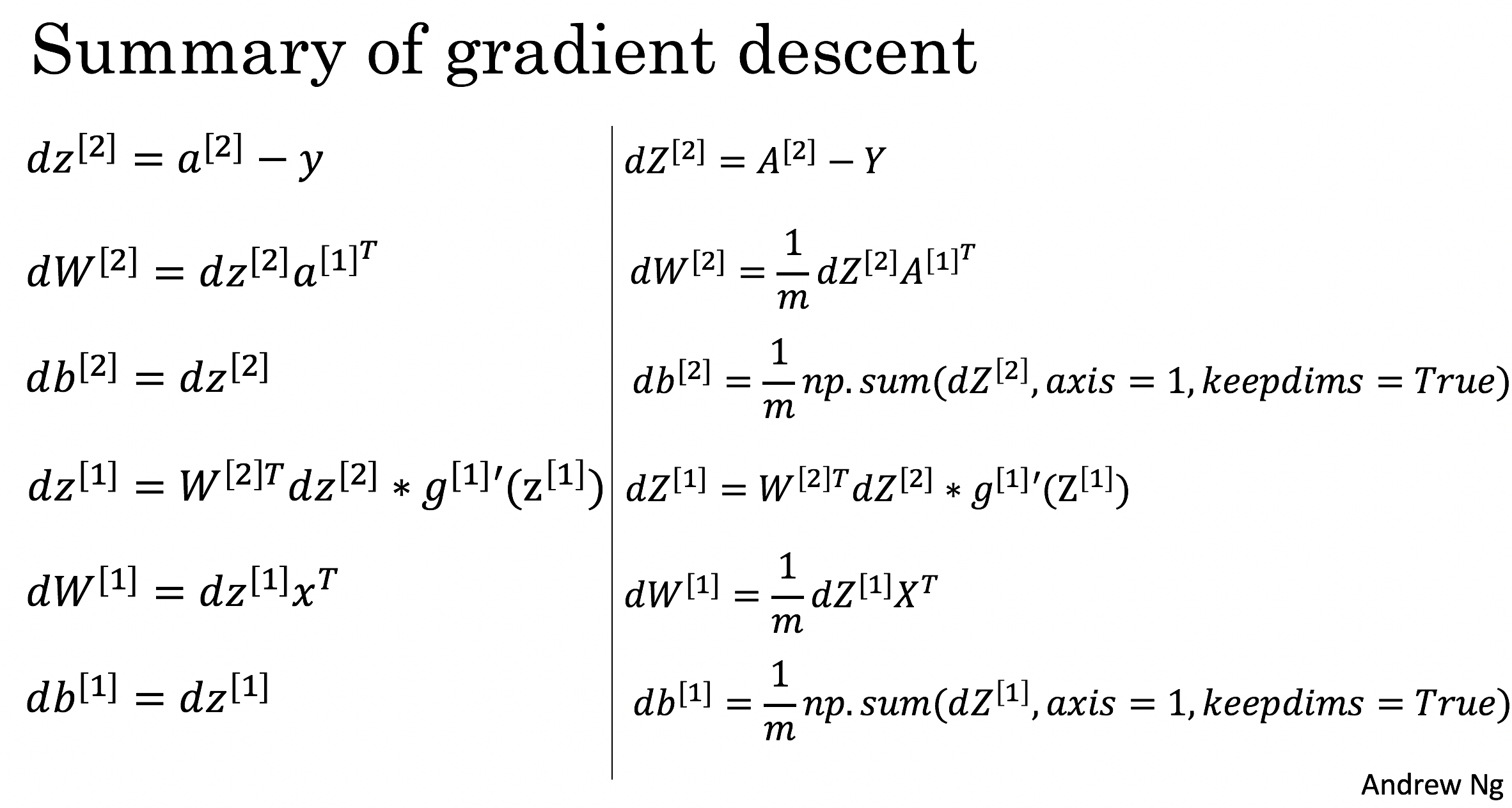
上图中左侧是单个实例的计算公式,右侧是m个实例矢量化后的计算公式。
随机初始化
当你开始训练神经网络时,将权重参数进行随机初始化非常重要。在逻辑回归的问题中,把权重参数初始化为零是可行的, 但把神经网络的权重参数全部初始化为零, 并使用梯度下降, 将无法获得预期的效果。
这里有两个输入样本参数,因此\(n^{[0]}\)等于2,还有两个隐藏单元,因此\(n^{[1]}\)也等于2。所以与隐藏层关联的权重\(w^{[1]}\) 是一个2x2的矩阵。现在我们将这个矩阵的初始值都设为0,同样我们将矩阵\(b^{[1]}\)的值也都初始化为零。偏差矩阵\(b^{[1]}\)的初始值都是0,不会影响最终结果。**但是将权重参数矩阵\(w^{[1]}\)初始值都设为零,会引起某些问题。这样的初始权重参数会导致,无论使用什么样的样本进行训练$ a^{[1]}_1\(与\)a^{[1]}_2\(始终是相同的,这第一个激活函数和这第二个激活函数将是完全一致的,因为这些隐藏神经元在进行完全相同的计算工作**。当你进行反向传播的计算时,由于 **对称问题**,这些隐藏单元将会在同样的条件下被初始化,最终导致\)z^{[1]}_1\(的导数和\)z^{[1]}_2\(的导数也不会有差别。同样的,假设输出的权重也是相同的,所以输出权重参数矩阵\)w^{[2]}\(也等于\)[0,0]$。但如果使用这种方法来初始化神经网络,那么上面这个隐藏单元和下面这个隐藏单元也是相同的,它们实现的是完全相同的功能,看可以称这是“对称”的。
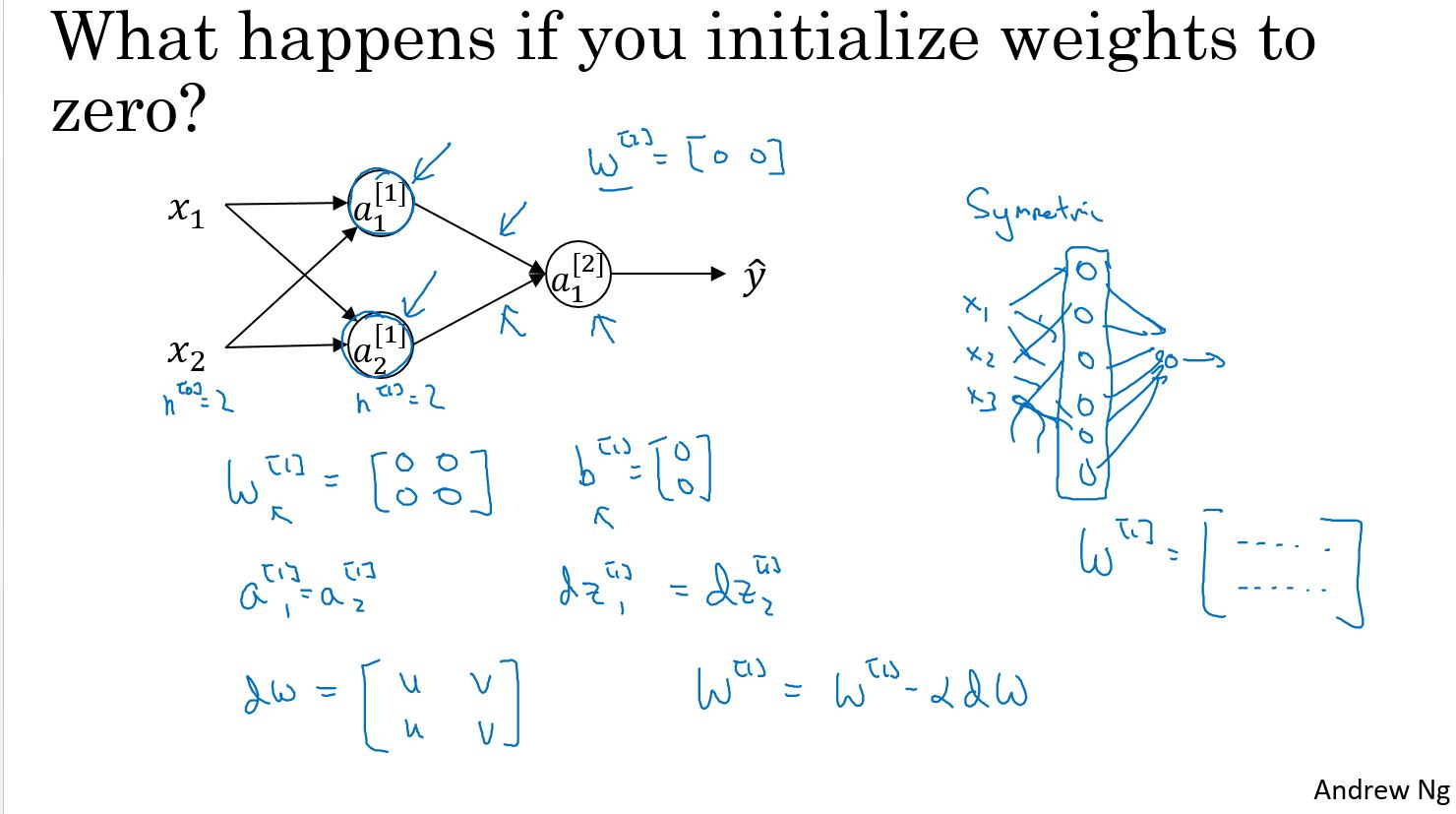
归纳一下这个结果:经过每一次训练迭代,将会得到两个实现完全相同功能的隐藏单元,\(W\)的导数将会是一个矩阵且每一行都是相同的,然后进行一次权重更新,当在实际操作时$ w^{[1]} \(将被更新成\) w^{[1]}-α*dw $时,将会发现,经过每一次迭代后 \(w^{[1]}\)的第一行与第二行是相同的。所以根据上述信息来归纳,可以得到一个证明结果:如果在神经网络中,将所有权重参数矩阵w的值初始化为零,由于两个隐藏单元肩负着相同的计算功能,并且也将同样的影响作用在输出神经元上,经过一次迭代后,依然会得到相同的结果。这两个隐藏神经元依然是“对称”的。同样推导下去,经过两次迭代 三次迭代,以及更多次迭代,无论将这个神经网络训练多久,这两个隐藏单元仍然在使用同样的功能进行运算。
编程练习
Planar data classification with one hidden layer
Welcome to your week 3 programming assignment. It's time to build your first neural network, which will have a hidden layer. You will see a big difference between this model and the one you implemented using logistic regression.
You will learn how to: - Implement a 2-class classification neural network with a single hidden layer - Use units with a non-linear activation function, such as tanh - Compute the cross entropy loss - Implement forward and backward propagation
1 - Packages
Let's first import all the packages that you will need during this assignment. - numpy is the fundamental package for scientific computing with Python. - sklearn provides simple and efficient tools for data mining and data analysis. - matplotlib is a library for plotting graphs in Python. - testCases provides some test examples to assess the correctness of your functions - planar_utils provide various useful functions used in this assignment
1 | # Package imports |
2 - Dataset
First, let's get the dataset you will work on. The following code will load a "flower" 2-class dataset into variables X and Y.
1 | X, Y = load_planar_dataset() |
Visualize the dataset using matplotlib. The data looks like a "flower" with some red (label y=0) and some blue (y=1) points. Your goal is to build a model to fit this data.
1 | # Visualize the data: |
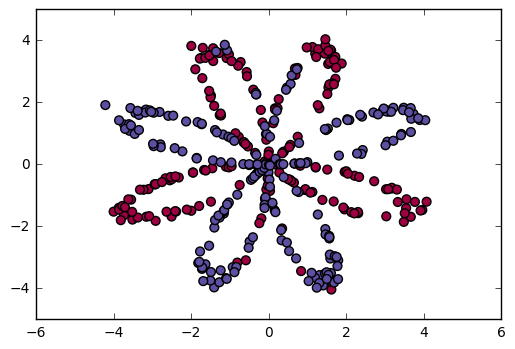
You have: - a numpy-array (matrix) X that contains your features (x1, x2) - a numpy-array (vector) Y that contains your labels (red:0, blue:1).
Lets first get a better sense of what our data is like.
Exercise: How many training examples do you have? In addition, what is the shape of the variables X and Y?
Hint: How do you get the shape of a numpy array? (help)
1 |
|
The shape of X is: (2, 400)
The shape of Y is: (1, 400)
I have m = 400 training examples!3 - Simple Logistic Regression
Before building a full neural network, lets first see how logistic regression performs on this problem. You can use sklearn's built-in functions to do that. Run the code below to train a logistic regression classifier on the dataset.
1 | # Train the logistic regression classifier |
You can now plot the decision boundary of these models. Run the code below.
1 | # Plot the decision boundary for logistic regression |
Accuracy of logistic regression: 47 % (percentage of correctly labelled datapoints)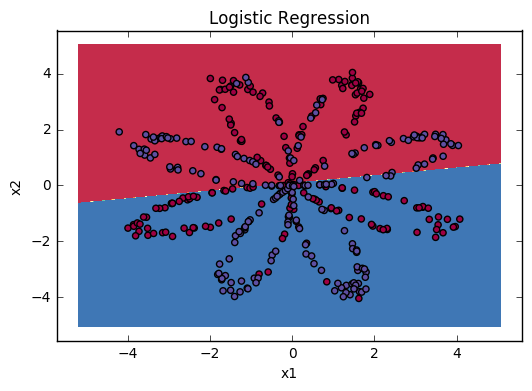
Interpretation: The dataset is not linearly separable, so logistic regression doesn't perform well. Hopefully a neural network will do better. Let's try this now!
4 - Neural Network model
Logistic regression did not work well on the "flower dataset". You are going to train a Neural Network with a single hidden layer.
Here is our model:
Mathematically:
For one example \(x^{(i)}\):
\[ z^{[1] (i)} = W^{[1]} x^{(i)} + b^{[1]} \]
\[ a^{[1] (i)} = \tanh(z^{[1] (i)}) \]
\[z^{[2] (i)} = W^{[2]} a^{[1] (i)} + b^{[2]} \]
\[ \hat{y}^{(i)} = a^{[2] (i)} = \sigma(z^{ [2] (i)}) \]
\[ y^{(i)}_{prediction} = \begin{cases} 1 & \text{if } a^{[2](i)} > 0.5 \\ 0 & \text{otherwise } \end{cases} \]
Given the predictions on all the examples, you can also compute the cost \(J\) as follows:
\[ J = - \frac{1}{m} \sum_{i=0}^{m} (y^{(i)}\log(a^{[2] (i)}) + (1-y^{(i)})\log(1- a^{[2] (i)})) \]
Reminder: The general methodology to build a Neural Network is to:
- Define the neural network structure ( # of input units, # of hidden units, etc).
- Initialize the model's parameters
- Loop:
- Implement forward propagation
- Compute loss
- Implement backward propagation to get the gradients
- Update parameters (gradient descent)
You often build helper functions to compute steps 1-3 and then merge them into one function we call nn_model(). Once you've built nn_model() and learnt the right parameters, you can make predictions on new data.
4.1 - Defining the neural network structure
Exercise: Define three variables:
- n_x: the size of the input layer
- n_h: the size of the hidden layer (set this to 4)
- n_y: the size of the output layer
Hint: Use shapes of X and Y to find n_x and n_y. Also, hard code the hidden layer size to be 4.
1 | GRADED FUNCTION: layer_sizes |
1 | X_assess, Y_assess = layer_sizes_test_case() |
The size of the input layer is: n_x = 5
The size of the hidden layer is: n_h = 4
The size of the output layer is: n_y = 24.2 - Initialize the model's parameters
Exercise: Implement the function initialize_parameters().
Instructions: - Make sure your parameters' sizes are right. Refer to the neural network figure above if needed. - You will initialize the weights matrices with random values. - Use: np.random.randn(a,b) * 0.01 to randomly initialize a matrix of shape (a,b). - You will initialize the bias vectors as zeros. - Use: np.zeros((a,b)) to initialize a matrix of shape (a,b) with zeros.
1 | # GRADED FUNCTION: initialize_parameters |
4.3 - The Loop
Question: Implement forward_propagation().
Instructions: - Look above at the mathematical representation of your classifier. - You can use the function sigmoid(). It is built-in (imported) in the notebook. - You can use the function np.tanh(). It is part of the numpy library. - The steps you have to implement are: 1. Retrieve each parameter from the dictionary "parameters" (which is the output of initialize_parameters()) by using parameters[".."]. 2. Implement Forward Propagation. Compute \(Z^{[1]}, A^{[1]}, Z^{[2]}\) and \(A^{[2]}\) (the vector of all your predictions on all the examples in the training set). - Values needed in the backpropagation are stored in "cache". The cache will be given as an input to the backpropagation function.
1 | # GRADED FUNCTION: forward_propagation |
Now that you have computed \(A^{[2]}\) (in the Python variable "A2"), which contains \(a^{[2](i)}\) for every example, you can compute the cost function as follows:
\[ J = - \frac{1}{m} \sum_{i = 0}^{m} ( y^{(i)}\log(a^{[2] (i)}) + (1-y^{(i)})\log(1- a^{[2] (i)} )) \]
Exercise: Implement compute_cost() to compute the value of the cost \(J\).
Instructions: - There are many ways to implement the cross-entropy loss. To help you, we give you how we would have implemented \[ \sum_{i=0}^{m} y^{(i)}\log(a^{[2](i)}) \]
1 | logprobs = np.multiply(np.log(A2),Y) |
(you can use either np.multiply() and then np.sum() or directly np.dot()).
1 | # GRADED FUNCTION: compute_cost |
Using the cache computed during forward propagation, you can now implement backward propagation.
Question: Implement the function backward_propagation().
Instructions: Backpropagation is usually the hardest (most mathematical) part in deep learning. To help you, here again is the slide from the lecture on backpropagation. You'll want to use the six equations on the right of this slide, since you are building a vectorized implementation.
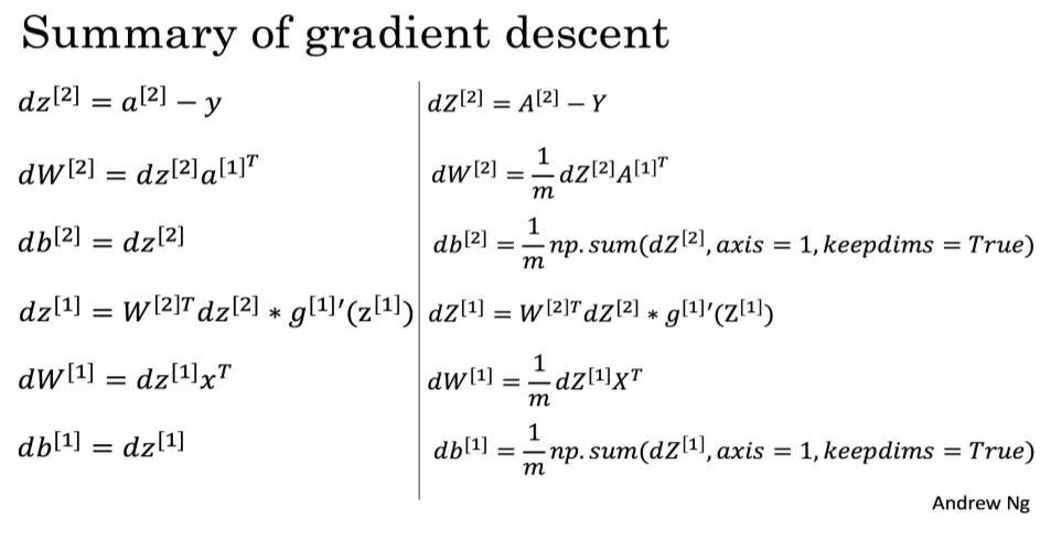
- Tips:
- To compute dZ1 you'll need to compute \(g^{[1]'}(Z^{[1]})\). Since \(g^{[1]}(.)\) is the tanh activation function, if \(a = g^{[1]}(z)\) then \(g^{[1]'}(z) = 1-a^2\). So you can compute \(g^{[1]'}(Z^{[1]})\) using
(1 - np.power(A1, 2)).
- To compute dZ1 you'll need to compute \(g^{[1]'}(Z^{[1]})\). Since \(g^{[1]}(.)\) is the tanh activation function, if \(a = g^{[1]}(z)\) then \(g^{[1]'}(z) = 1-a^2\). So you can compute \(g^{[1]'}(Z^{[1]})\) using
1 | # GRADED FUNCTION: backward_propagation |
Question: Implement the update rule. Use gradient descent. You have to use \((dW1, db1, dW2, db2)\) in order to update \((W1, b1, W2, b2)\).
General gradient descent rule: $ = - $ where \(\alpha\) is the learning rate and \(\theta\) represents a parameter.
Illustration: The gradient descent algorithm with a good learning rate (converging) and a bad learning rate (diverging). Images courtesy of Adam Harley.
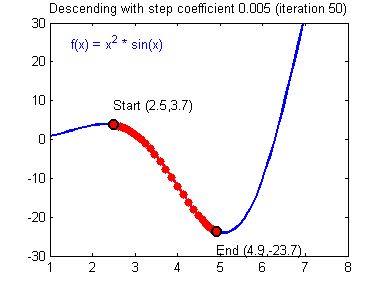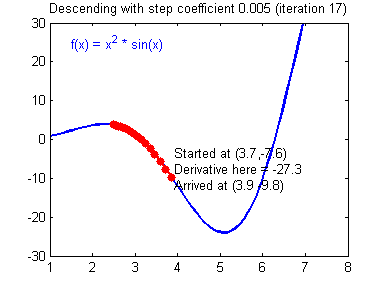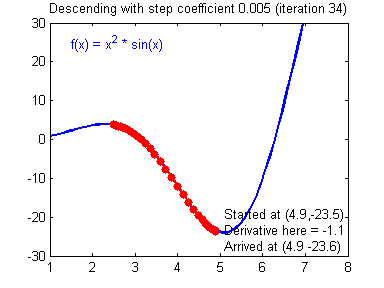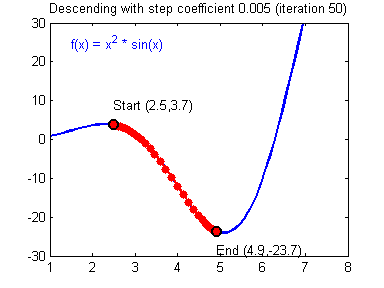
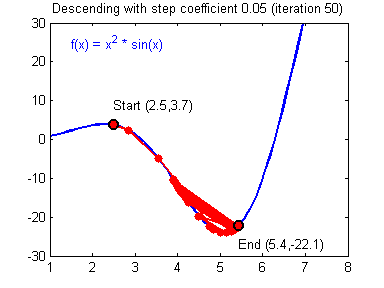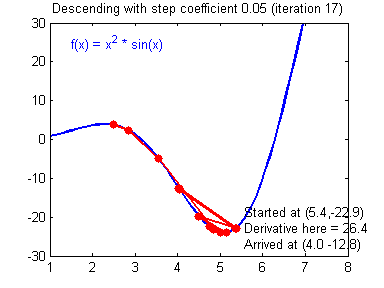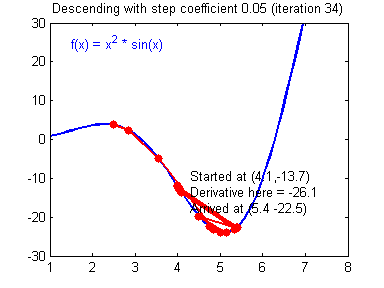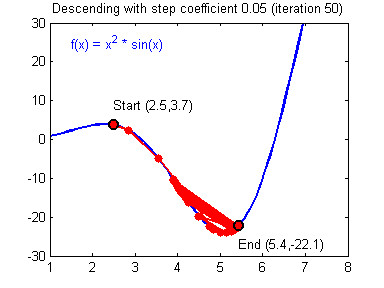
1 | # GRADED FUNCTION: update_parameters |
4.4 - Integrate parts 4.1, 4.2 and 4.3 in nn_model()
Question: Build your neural network model in nn_model().
Instructions: The neural network model has to use the previous functions in the right order.
1 | GRADED FUNCTION: nn_model |
1 | X_assess, Y_assess = nn_model_test_case() |
Cost after iteration 0: 0.692739
Cost after iteration 1000: 0.000218
Cost after iteration 2000: 0.000107
Cost after iteration 3000: 0.000071
Cost after iteration 4000: 0.000053
Cost after iteration 5000: 0.000042
Cost after iteration 6000: 0.000035
Cost after iteration 7000: 0.000030
Cost after iteration 8000: 0.000026
Cost after iteration 9000: 0.000023
W1 = [[-0.65848169 1.21866811]
[-0.76204273 1.39377573]
[ 0.5792005 -1.10397703]
[ 0.76773391 -1.41477129]]
b1 = [[ 0.287592 ]
[ 0.3511264 ]
[-0.2431246 ]
[-0.35772805]]
W2 = [[-2.45566237 -3.27042274 2.00784958 3.36773273]]
b2 = [[ 0.20459656]]4.5 Predictions
Question: Use your model to predict by building predict(). Use forward propagation to predict results.
Reminder: \[ predictions = y_{prediction} = \begin{cases} 1 & \text{if}\ activation > 0.5 \\ 0 & \text{otherwise} \end{cases} \]
As an example, if you would like to set the entries of a matrix X to 0 and 1 based on a threshold you would do: X_new = (X > threshold)
1 | # GRADED FUNCTION: predict |
1 | parameters, X_assess = predict_test_case() |
predictions mean = 0.666666666667It is time to run the model and see how it performs on a planar dataset. Run the following code to test your model with a single hidden layer of \(n_h\) hidden units.
1 | # Build a model with a n_h-dimensional hidden layer |
1 | Cost after iteration 0: 0.693048 |
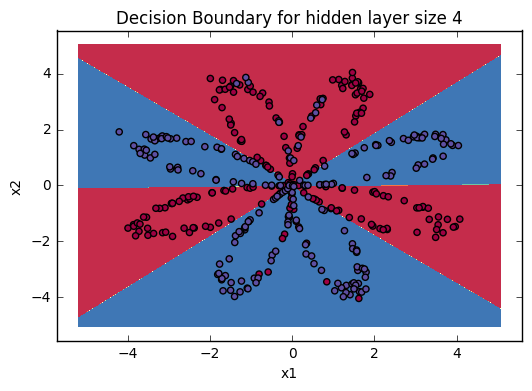
Accuracy is really high compared to Logistic Regression. The model has learnt the leaf patterns of the flower! Neural networks are able to learn even highly non-linear decision boundaries, unlike logistic regression.
Now, let's try out several hidden layer sizes.
4.6 - Tuning hidden layer size (optional/ungraded exercise)
Run the following code. It may take 1-2 minutes. You will observe different behaviors of the model for various hidden layer sizes.
1 | # This may take about 2 minutes to run |
Accuracy for 1 hidden units: 67.5 %
Accuracy for 2 hidden units: 67.25 %
Accuracy for 3 hidden units: 90.75 %
Accuracy for 4 hidden units: 90.5 %
Accuracy for 5 hidden units: 91.25 %
Accuracy for 20 hidden units: 90.0 %
Accuracy for 50 hidden units: 90.25 %
Interpretation: - The larger models (with more hidden units) are able to fit the training set better, until eventually the largest models overfit the data. - The best hidden layer size seems to be around n_h = 5. Indeed, a value around here seems to fits the data well without also incurring noticable overfitting. - You will also learn later about regularization, which lets you use very large models (such as n_h = 50) without much overfitting.
Optional questions:
Note: Remember to submit the assignment but clicking the blue "Submit Assignment" button at the upper-right.
Some optional/ungraded questions that you can explore if you wish: - What happens when you change the tanh activation for a sigmoid activation or a ReLU activation? - Play with the learning_rate. What happens? - What if we change the dataset? (See part 5 below!)
You've learnt to: - Build a complete neural network with a hidden layer - Make a good use of a non-linear unit - Implemented forward propagation and backpropagation, and trained a neural network - See the impact of varying the hidden layer size, including overfitting.
Nice work!
5) Performance on other datasets
If you want, you can rerun the whole notebook (minus the dataset part) for each of the following datasets.
1 | # Datasets |
Congrats on finishing this Programming Assignment!
Reference: - http://scs.ryerson.ca/~aharley/neural-networks/ - http://cs231n.github.io/neural-networks-case-study/
小结
本周的课程包含: - 含有单独隐藏层的神经网络 - 参数初始化 - 使用前向传播进行预测 - 以及在梯度下降时使用反向传播中 - 涉及的导数计算